COVID-19 Data Workflow
🏆 I received the 2020 NIAID Merit Award from the office of Dr. Anthony Fauci for my work on this project!
Overview
Background
As the COVID-19 pandemic raged on, hospitals around the world generated massive amounts of clinical patient data. The National Institutes of Health (NIH) received these international datasets for lifesaving research purposes in sporadic, non-standard deliveries. Initially, our team of bioinformaticians and developers set up ad-hoc data processing solutions.
Clinicians and researchers were scrambling to get their hands on the valuable data, to help diagnose patients and understand the inflammatory disorders that COVID-19 causes. The influx of data necessitated a standardized data processing pipeline, and a central source to access and analyze it.
Role
Member of the COVID Data Management Task Force under Dr. Fauci:
liaison between developers, analysts, and clinicians
developer and data analyst
data sprint team lead
Goal
Craft a seamless clinical data access and analysis experience, minimizing manual human error, and enabling healthcare practitioners to focus on lifesaving COVID-19 research.
Solution + Impact
Developed an automated data cleaning, validation, upload, and visualization pipeline. Eliminated human error, democratized data analysis capabilities, and increased data freshness.
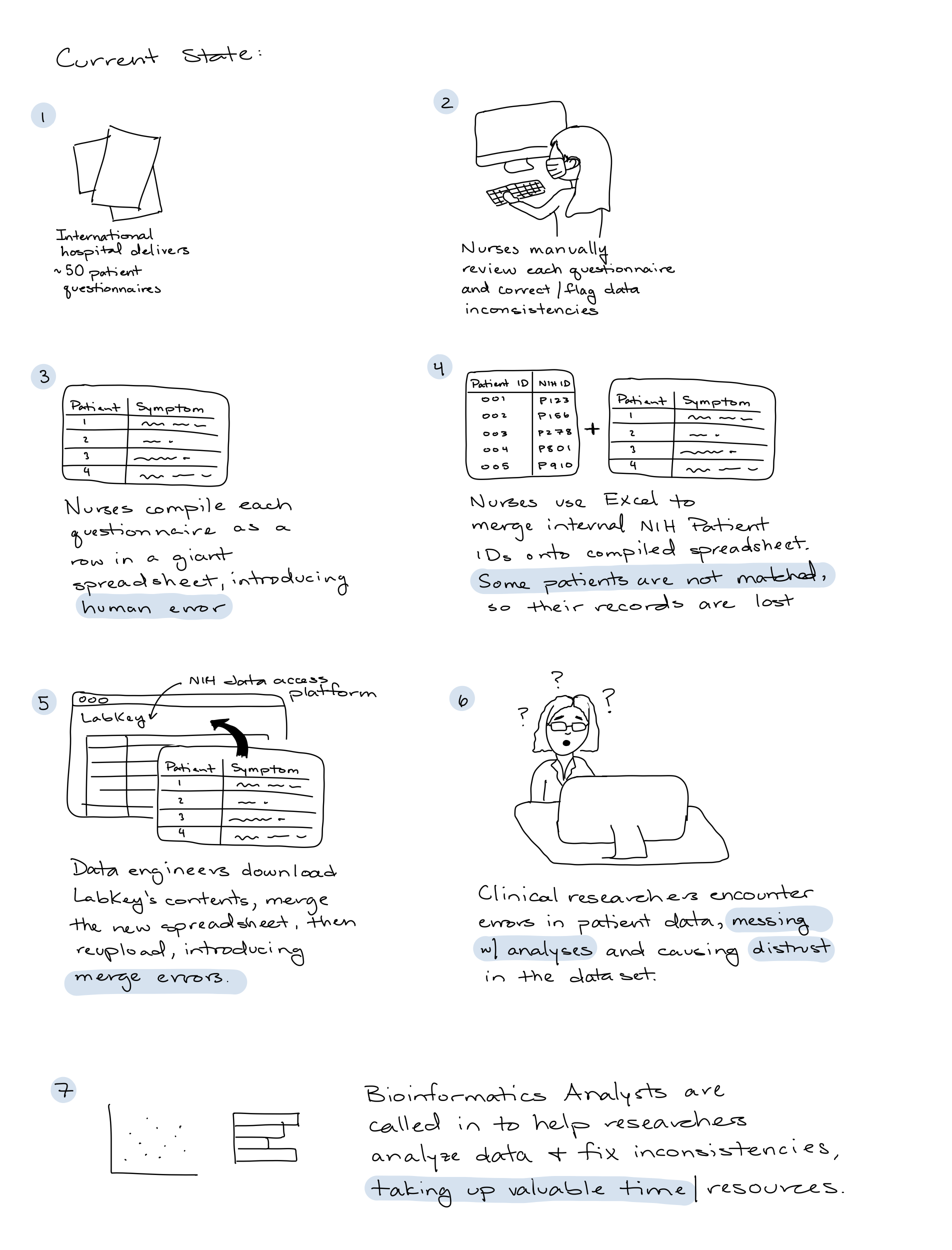
Current state
Large data deliveries of individual patient questionnaires were manually parsed, and combined into a single Excel spreadsheet by nurses.
To upload to the central data access platform, LabKey, data engineers downloaded all of the contents, appended the new Excel spreadsheet, and re-uploaded.
The end customer, clinical researchers, would download patient data from LabKey and encounter data errors, causing them to mistrust the dataset.
Bioinformatics Analysts were recruited by researchers to make sense of the messy data, and generate visualizations and preliminary analyses.
Goal Setting
After speaking with the nurses, data engineers, researchers, and bioinformatics analysts, I synthesized a list of goals for our team to accomplish through coding sprints:
Decrease manual parsing time for each batch
Flag inconsistencies + reduce human error
Develop modular, re-runnable solution
Automate LabKey upload process
Part 1: Aggregate + Clean
Stakeholder Needs
Nurses: "We're spending valuable time away from patients on data deliveries. It's hard to match patients with NIH IDs get catch all the data errors"
Data Engineers: "The data delivery we receive should be well organized, free from errors, and maintain processed + raw versions of the data"
Researchers: "We don't trust the data in LabKey. Can you provide us the underlying raw data?"
Process
NIAID decided on a standardized questionnaire to capture clinical data for one patient, over time. Each questionnaire had 9 data types, a patient ID, and a date. One patient could have multiple questionnaires, for data captured on them over several days.
The data folks needed the data split by type, in some consistent, aggregated format. I wrote a parsing script in Python that aggregated each questionnaire into a single Excel spreadsheet, with tabs for each type of data. This file was both human readable, and machine ingestible.
Solution
Re-runnable script to automatically clean, flag, and compile questionnaire data delivery.
Input: folder of patient questionnaires
Output: Excel spreadsheet compilation
Flagged data inconsistencies
Raw data alongside cleaned data
Automatically matched NIH patient ID
Part 2: Human Review
Stakeholder Needs
Nurses: "There are certain clinical signatures that only we can catch, but we can't sift through every patient file. We need automation + human review for edge cases"
Data Engineers: "We need to automate the data upload process, appending new deliveries to the old. The manual process takes too long!"
Process
I interviewed nurses and did A/B testing to understand which format of compiled questionnaires was easiest for them to sort through. I experimented with warning messages and ways of flagging inconsistencies.
I worked with a few data engineers to develop a pipeline that automatically scans a designated folder in the NIH server for review deliveries. Overnight, a script ingests the data and appends it to the existing data in LabKey. This development took several rounds to perfect, due to security and processing roadblocks.
Solution
The output of the script from Part I was human and machine readable, meaning data engineers could easily upload it, while nurses could easily parse through and edit inconsistencies.
Once nurses were done editing, they could drag and drop the spreadsheet into a designated folder, and the delivery would automatically show up in LabKey the next day!
Part 3: Explore + Analyze
Stakeholder Needs
Researchers: "The lab value data in LabKey is difficult to analyze. Analysts take a while. We want simple data queries answered fast."
Bioinformatics Analysts: "Our client queue is ever growing. We need to give researchers some self service analysis capabilities"
Process
A hole that remained in this processing pipeline was that clinicians and researchers had to download the huge datasets, open it in Excel, and try generating some rudimentary analyses and visualizations.
To save my bioinformatics analysts some time, I worked with a team that was building live analytics and visualization tools in the cloud. This database (we'll call it Cloud Analytics Tool) separately stored laboratory data on the same patients that were in LabKey.
We identified an opportunity to connect the two services. I developed an automated process that matched records across the two databases using the unifying NIH ID. For each record in LabKey, the end users could click straight into the Cloud Analytics Tool and enjoy the integrated visualization and analysis capabilities
Solution
Connect the Laboratory Values dataset in LabKey with the Cloud Analytics Tool, enabling clinical researchers to answer questions on the dataset themselves, rather than waiting on bioinformatics analysts.
Impact
The process from questionnaire delivery to upload went down from 2-3 weeks to 1-3 days.
Nurses avoided hours of manual labor editing/compiling questionnaires
Data Engineers enjoyed automatic + streamlined upload
Clinical Researchers had access to fresh + error free data
Bioinformatics Analysts had more time to focus on deeper analyses rather than data cleaning and preliminary data exploration.
Future Improvements
Many nurses weren’t well versed with Terminal or the command line, so even typing out a simple command was difficult. Given more time, I’d like to implement a Graphical User Interface with a simple button to point the software to the delivery folder, and output the processed data delivery.
Lessons Learned
Leading a development team takes patience, perseverance, and a unifying vision
A solution that benefits the end customer (the clinical researcher) necessitates teared mini-solutions for internal stakeholders that add up to a smooth overall experience.